Introduction
Skeletal muscle regeneration is a complex process where various cell types and cytokines are involved. Single-cell RNA-sequencing (scRNA-seq) provides the opportunity to deconvolute heterogeneous tissue into individual cells based on their transcriptomic profiles. Recent scRNA-seq studies on mouse muscle regeneration have provided insights to understand the transcriptional dynamics that underpin muscle regeneration. However, a database to investigate gene expression profiling during skeletal muscle regeneration at the single-cell level is a deficiency. Here, we collected over 105,000 cells at 7 key regenerative time-points and non-injured muscles and developed a database, the Single-cell Skeletal Muscle Regeneration Database (SCSMRD). SCSMRD allows users to search the dynamic expression profiles of the interested genes across different cell types during the skeletal muscle regeneration process. It also provides a network to show the activity of regulons in different cell types at different time points. Pesudotime analysis showed the state changes trajectory of muscle stem cells (MuSCs) during skeletal muscle regeneration.
Search functions
To provide users easy access to the SCSMRD, we designed auser-friendly interface to allow users to perform various op-erations. We provided four main functionalities: (1) Gene ex-pression level search in different cell populations along withdifferent time points, (2) Cell-type-specific regulon networksearch, (3) Dynamic gene expression changes along with thecontinuous myogenic cell-states search.
Gene expression level search
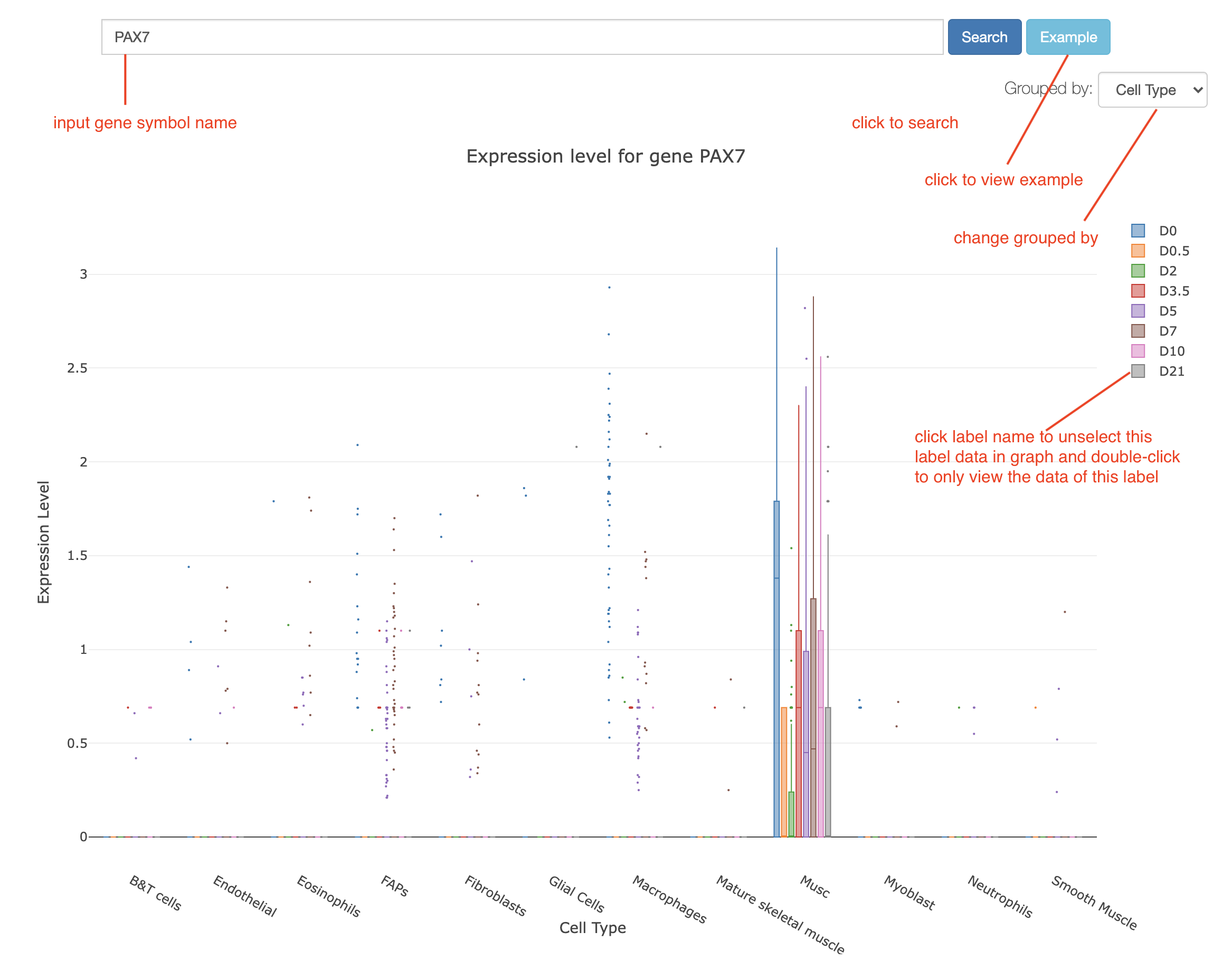
For the gene expression level search, our database allows users to visualize the search result with an interactive grouped Boxplot (Figure 1). Users can view the gene expression level of an interesting gene in different cell clusters by giving a gene symbol (e.g., PAX7) in the search input box. Specifically, users can click the "Example" button to view the default gene expression level in SCSMRD. Furthermore, an interactive Boxplot of gene expression level grouped by different cell clusters (e.g. cell type or cell date) is displayed by clicking the "Search" button. Users can select different grouping rules on the menu in the upper left corner to switch the Boxplot between different clustering rules. Clicking on the cluster name listed in the graph legend, (e.g. "D0" in Figure 1) can remove the Boxplot data of this cluster. Moreover, double-clicking on a cluster name allows users to view the detailed gene expression value of this cluster. These interactive functions allow users to compare gene expression levels only for their interesting clusters. Finally, the Boxplot can be downloaded in PNG format for further usage.
Regulon netwrok search
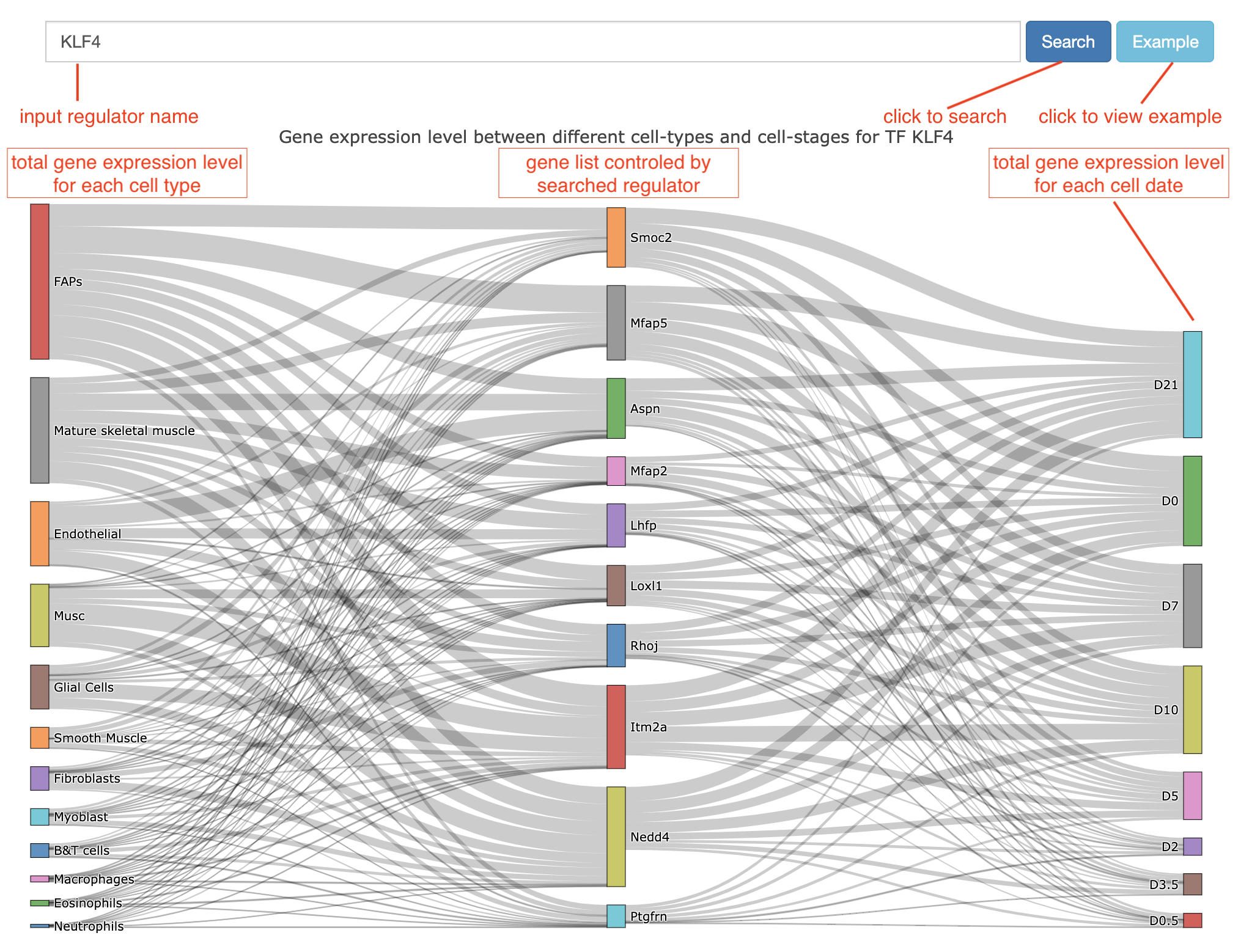
For cell-type-specific regulon search, we designed an interactive Sankey diagram to present the cell-type-specific regulon network(Figure 2). Users can search interested regulator factors by inputting the gene symbol in the search input box. As demonstrated in Figure 2, the result Sankey diagram is composed of three columns that are connected by lines. The middle column of the Sankey diagram is a group of genes controlled by the searched regulator. The left and right columns connected to the middle column are the total expression level of cells grouped by different cell types and different cell time points, respectively. Specifically, users can drag the items in each column to reorder the Sankey graph(e.g. reordering the column according to the total expression level). Also, the expression level is reflected by the thickness of the connected lines and the detailed expression value can be viewed by moving the cursor on the specific line.
Cell pseudotime search
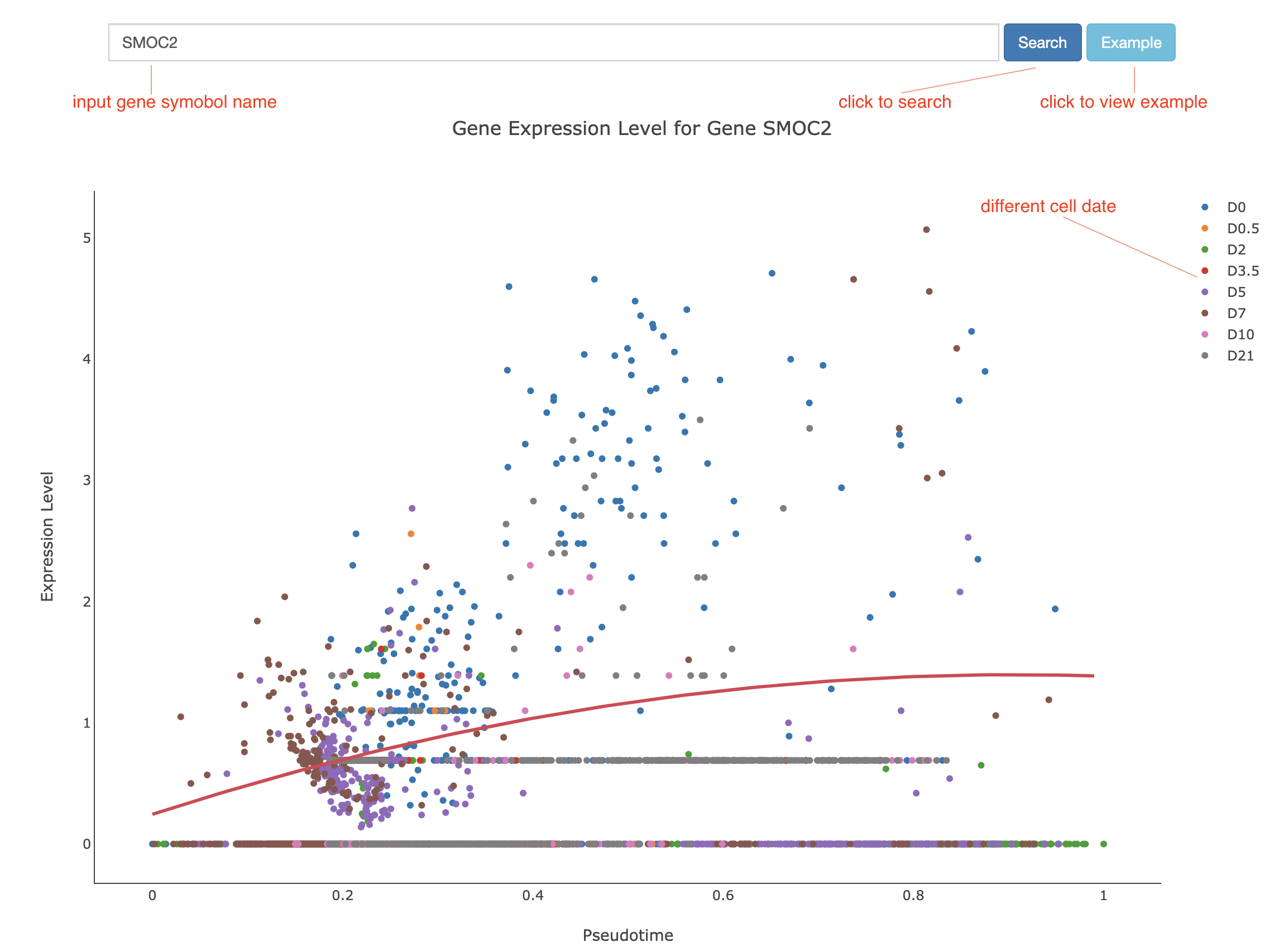
For cell pseudotime search, we represented the expression level changes of genes along the continuous myogenic cell-states with an interactive Scatter plot appending with the linear regression line(Figure 3). Users can view the gene expression changes in muscle cells along with pseudotime trajectory.
Download
User can download the data resources of SCSMRD from our download page.